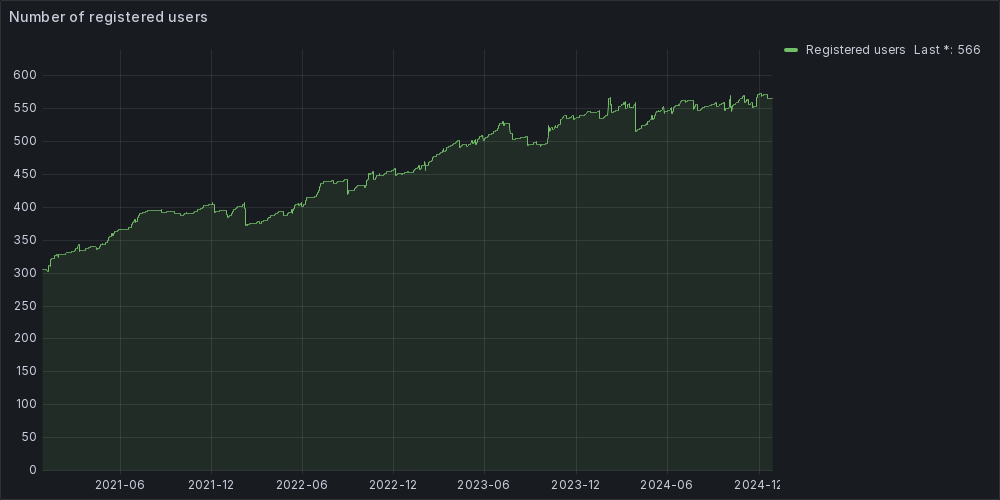
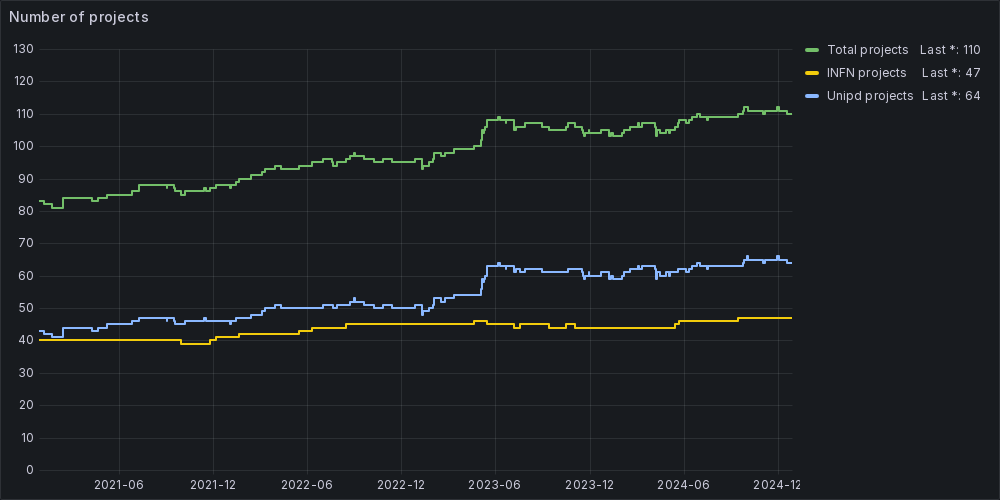
Currently, we provide support to about 600 users involved in more than 110 research projects led by the communities of INFN and the University of Padua. The trend indicates that the number of users and projects grows over time.
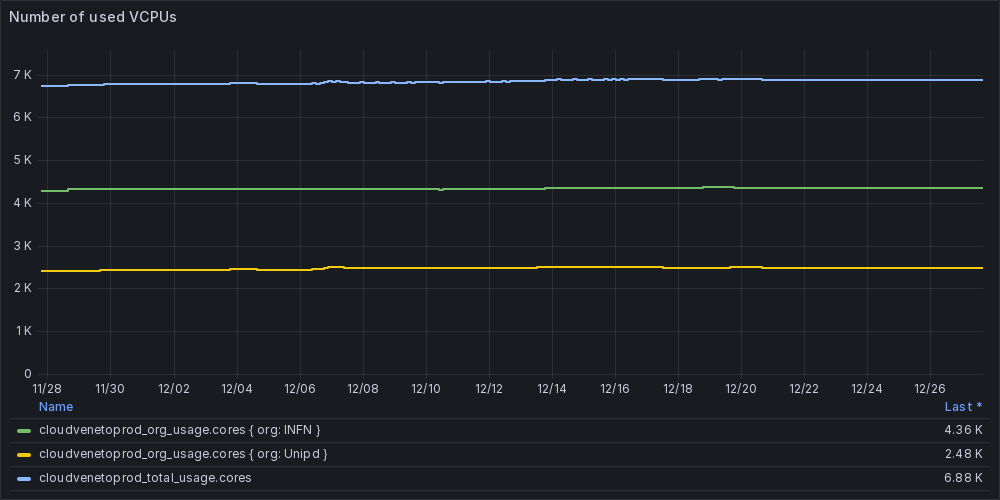
Resource usage in the last month

As an indication of the usage of CloudVeneto, we highlight brief descriptions of some of the projects.

We use relativistic Density Functional Theory to investigate the NMR properties of organometallic complexes of heavy atoms. In Figure we show two complexes of Osmium exhibiting opposite effects in the chemical shift d(187Os), due to the spin-orbit (SO) coupling in the Hamiltonian: without the inclusion of the SO contribution the calculated values are wrong by several hundreds of ppm, while inclusion of the SO term results in a nice agreement with the experiments.
Giacomo Saielli (XeDFTNMR)

The project would like to verify the possibility of exciting plasmons in the UV spectral region (below 400 nm) for metals which are particularly stable in aggressive environments. The UV spectral region is interesting for detection of biological molecules. An example of a BEM calculation of the enhancement factors for a Pt nanoparticle of 20 nm of diameter is reported in the front panel image.
Moreno Meneghetti (Plasmonics)

The NanoStructures Group (NSG) at DFA within the NanoEM project of CloudVeneto investigates the light-matter interaction at the nanoscale in the visible and infrared spectral region employing computational-intensive numerical methods. The aim is the development of innovative coherent or single-photon light sources and of more efficient nonlinear optical devices.
Giovanni Mattei (NanoEM)

The Padua group is actively involved in data management for the upcoming CTAO project. Robust storage solutions are essential given the expected data volume of hundreds of petabytes per year. CloudVeneto resources are used as a test-bed for deploying and validating the dCache storage element. This distributed storage infrastructure is implemented and tested within CloudVeneto, ensuring system reliability and supporting future upgrades.
Davide Miceli (MAGIC_CTA)

In nuclear astrophysics, determining nuclear reaction cross sections is key to understanding nucleosynthesis and stellar evolution. Using R-matrix theory, we compared frequentist and Bayesian approaches to estimate these cross sections and their uncertainties. The analysis assessed various statistical techniques, highlighting the role of proper systematic uncertainty treatment and covariance matrix estimation to enhance reliability and reproducibility.
Jakub Skowronski (LUNA)

This study evaluates machine learning techniques to determine the most effective feature selection method for regression modeling in omics data. Through simulation, elastic net emerged as the best approach for selecting relevant variables. However, computational resource optimization remains a challenge for large datasets, necessitating further methodological advancements.
Luca Vedovelli (Feature Selection)

The project consists of a series of simulations aimed at studying the role of raters' thresholds in estimating interrater agreement. Two Bayesian models are developed to mimic the process of Boolean classification in humans. Several scenarios are tested through an extensive Monte Carlo simulation to evaluate the accuracy of the new methodology for estimating raters' thresholds and the actual degree of agreement among independent raters.
Massimo Nucci (DPG - Data Analysis and Simulations)

We derive bounds on the dark matter (DM) lifetime using prospective measurements of the 21-cm monopole signal. First, we update the bounds for the single-component DM scenario, employing state-of-the-art modeling of energy deposition in the intergalactic medium. We then explore two additional possibilities: cold DM with a decaying sub-component and a nearly degenerate two-component DM model.
Francesco D’Eramo (Dark Matter Bounds from 21-cm Cosmology)

This project concerns the comparison of different methods for authorship attribution. The techniques used mainly involve a machine learning (ML) framework and the use of Large Language Models by fine-tuning them. ML procedures use a variety of vector representations of text, including calculation of specific indices, word frequency selection, and dimensionality reduction techniques. Applications mainly concern narrative and AI detection.
Andrea Sciandra (auth_attr)

The ALICE experiment at CERN uses collisions of lead nuclei to study the quark-gluon plasma phase of matter which characterized the Universe after the Big Bang. The Padua ALICE team is studying its influence on the energy spectrum of particles containing charm and beauty quarks, which are rare and decay before reaching the detector. Cloud Veneto is used for exploiting Machine Learning for signal-background separation.
Andrea Rossi (ALICE)

Cherenkov telescope images of cosmic and gamma ray showers are ideal for AI analysis to classify events, estimate gamma ray energy, and determine direction. Deep learning, especially convolutional neural networks, is being explored for its ability to detect rare events, outperforming traditional methods in identifying challenging cases like multiple gamma rays or heavy nuclei.
Dr. Rubèn Lopez Coto

The CMS experiment at CERN is searching for new particles like the Higgs boson and measuring particle-antiparticle asymmetries. The Padua CMS group is involved in these studies, analyzing large data sets from the detector. CloudVeneto's computing resources are used for data reconstruction, simulations, statistical analysis, and machine learning models to distinguish signals from background processes.
Dr. Jacopo Pazzini

This project, in collaboration with the E. Mach Institute, analyzed sequence similarity in a database of 300,000 plant protein sequences from commercially important plants like apple, strawberry, and coffee. Self-alignment studies were conducted to cluster biologically significant sequences. CloudVeneto's platform was used to parallelize the task, reducing computation time.
Dr. Ivan Mičetić

The project on the CloudVeneto platform simulates the recognition process between chemical compounds and target proteins using a molecular docking approach. Our laboratory (MMS) has archived a public chemical library of around 5 million compounds for drug candidate screening (MMsINC). Each molecular docking simulation generates approximately 5 plausible ligand-protein complexes per target protein, resulting in around 25 million complexes per study.
Prof. Stefano Moro

The activities of the group are focused on developing and exploiting numerical and theoretical methods for investigating quantum many-body systems, bridging them to novel strategies for investigating open problems in condensed matter, high energy physics and quantum science and technologies. We develop the Quantum Tea tensor network library, a general purpose tensor network software suite.
Simone Notarinicola (QST)

The project explores nuclear theory in advancing medical applications, focusing on the production of theranostic radionuclides like 47Sc and 155Tb. Through nuclear science integration, it improves precision treatment and patient outcomes. Key results include a new patent for 47Sc production and a target optimization for 155Tb.
Luciano Canton (RIMA)

Small proteins usually follow two-state folding kinetics with no intermediates. The presence of entangled motifs in an antifreeze protein structure was shown to result in more complex behavior within coarse-grained simulations. Two distinct kinetic intermediates are populated: a short-lived entangled state conducive to fast folding and a longer-lived non-entangled state, acting as a kinetic trap.
Antonio Trovato (PhaseSepProt)

The Large-Sized Telescope, the first telescope of the CTAO project, is nearing the completion of its commissioning phase, with the first scientific data already collected. The Padua group is deeply involved in these activities. CloudVeneto resources are used to refine and upgrade the telescope pointing system. Additionally, the analysis of scientific data will enable studies in fundamental physics at the frontier of research
Davide Miceli (MAGIC_CTA)

LUXE at DESY explores non-linear QED by colliding 16.5 GeV electrons with a 40 TW laser. A key step in reconstructing the process is inferring laser intensity from the gamma angular distribution. Simulations in CloudVeneto were crucial to assess this method, estimate the signal in a realistic setup, and predict charge collection in sapphire, enabling a novel analysis of experimental data.
Pietro Grutta (LUXE)

Spatial metapopulation models study how landscape structure influences global species dynamics. We extend this framework to finite populations, analyzing how limited local habitat capacities and stochasticity impact species persistence. By deriving the finite-size scaling of survival probability and extinction time moments, we characterize the critical transition between survival and extinction.
Alice Doimo (Ecological_simulations)

The purpose of the project is to provide forecast estimates for the incidence at the national and regional level of seasonal influenza in Italy. This is achieved by calibrating a metapopulation compartmental epidemic model with mobility of individuals on the data provided by the influcast.org platform. Our estimates are then aggregated to produce a more precise ensemble model.
Manlio De Domenico (DARECloud)

Networks span every scale imaginable. At CoMuNe lab, we study networks on every scale, ranging from microscopic metabolic networks (left), to ecological meta-population models (middle), to building global risk maps related to extreme climatic events (right). The Cloud Veneto platform enables us to perform extensive simulations and data analyses to study the interplay between scales.
Manlio De Domenico (Complex Multilayer Networks)

Controlling the interaction between plasma and surfaces in atmospheric pressure plasma jets is still challenging since the propagation is governed by instabilities. Key factors include electron density and temperature, induced electric fields, current, and localized heating. Thanks to 2D modeling we showed the possibility to obtain a diffused plasma on the surfaces and to control the ion flux.
Alessandro Patelli (APPJ)

CATHY (CATchment HYdrology) is an integrated surface-subsurface hydrological model for the simulation of complex hydrological processes to improve our understanding of watershed hydrology and surface water – groundwater interactions. It also provides data assimilation capabilities to incorporate information from measured data directly into the model and to quantify uncertainty. CloudVeneto is crucial for running the model in the most demanding test cases.
Matteo Camporese (HydrologicalDA)

The Padua group, involved in building components for the Large-Sized Telescopes of CTA project, is refining the data analysis software. Using CloudVeneto resources, tools for event reconstruction are being developed, simulating how the telescope observes atmospheric showers and defining methods to determine the direction and energy of gamma rays. CloudVeneto will be used to analyze the first scientific data from the telescope inaugurated in La Palma.
Dr. Rubèn Lopez Coto

The CMS experiment at CERN produces tens of PBs of data annually. The Padua CMS group aims to redefine high-energy physics computing by integrating modern Big Data technologies. Additionally, innovative real-time data acquisition techniques using fast data streaming systems based on Apache Kafka are developed. CloudVeneto provides dedicated clusters for these activities, enhancing data analysis and acquisition efficiency.
Dr. Jacopo Pazzini

The QuantumFuture group at DEI has utilized CloudVeneto for a project involving a quantum random number generator with data rates of tens of Gbps. On CloudVeneto, post-processing and analysis of data from the physical generator were performed. We appreciated having total control over the resources and the ability to dynamically manage the number of machines and resources.
Dr. Marco Avesani

The Computational Biology Lab of the Department of Biomedical Sciences maintains a database of structural annotations for disordered regions in protein sequences, originally covering 80 million sequences. An update is underway to recalculate annotations for over 130 million sequences. CloudVeneto's platform is used to parallelize the annotation pipeline, reducing computation time and speeding up the release cycle.
Dr. Ivan Mičetić

Antisymmetrized Molecular Dynamics (AMD) is a code used to simulate nuclear reaction dynamics, incorporating particle structures and correlations. The NUCLEX collaboration utilizes the CloudVeneto infrastructure, running AMD in a virtual machine cluster with parallel processing via OpenMPI. This setup reduces computation time from several months to just 5-7 days for 50,000 events.
Dr. Tommaso Marchi, Dr. Magda Cicerchia
Currently, we provide support to about 550 users involved in more than 100 research projects led by the communities of INFN and the University of Padua.

Currently, we provide support to about 550 users involved in more than 100 research projects led by the communities of INFN and the University of Padua.

The trend indicates that the number of users and projects grows over time.

Some information about the current VCPUs usage
The images of atmospheric showers produced by cosmic rays and gamma rays captured by Cherenkov telescopes are well-suited for analysis by artificial intelligence for the purpose of:
Several deep learning methodologies are under study, particularly convolutional neural networks trained on simulated samples, whose performance will be compared with more traditional analysis methods. The advantage of the “deep learning” approach is the ability to conduct searches for rare and specific events such as multiple gamma rays (bosonic condensates) or images of showers produced by “heavy nuclei” in cosmic rays, which are difficult to classify with analytical methods.
Dr. Rubèn Lopez Coto
Dr. Rubèn Lopez Coto
The CMS (Compact Muon Solenoid) experiment focuses on detecting particles produced in proton collisions generated by the Large Hadron Collider (LHC) at CERN, with the aim of conducting an extensive campaign of measurements. Among these, notable examples include searches for ‘new’ particles, such as the discovery of the Higgs boson, or precision measurements of fundamental properties of nature, as in the case of measuring particle-antiparticle asymmetries.
The CMS group at the Department of Physics and Astronomy of Padua and the INFN section of Padua is actively involved in many of these studies, based on the analysis of vast amounts of data collected by the detector’s sensors.
For the reconstruction and subsequent analysis of this data, the use of computing tools capable of tackling computationally intensive tasks is necessary. These tasks include simulating numerous interactions between particles and the detector’s response, reconstructing events with highly complex topologies, performing statistical analysis for estimating confidence intervals, or developing classifiers based on neural networks for discriminating possible signals from the expected background from known processes in the detector.
To this end, the resources provided by the CloudVeneto infrastructure through dedicated clusters are extensively utilized, including the creation of clusters with ‘elastic’ resource allocation, for the utilization of the experiment’s reconstruction software (CMSSW, based on Python and C++) and development environments for multivariate algorithms, such as TMVA, TensorFlow, and Theano.
Dr. Jacopo Pazzini
The CMS (Compact Muon Solenoid) experiment at CERN studies the outcomes of proton-proton collisions produced by the Large Hadron Collider: having to deal with the analysis of signals from over 70 million detector readout channels every 25 ns, tens of PB of data are produced annually. Analyzing these large datasets to search for rare and highly elusive signals requires intensive use of extensive computing resources, both for the selection and processing of data collected ‘online’ during acquisition, and in the subsequent ‘offline’ data analysis.
The CMS group at the Department of Physics and Astronomy of Padua, and the INFN section of Padua collaborate with CERN with the aim of redefining the computing paradigm in high-energy physics analyses through the integration of modern technologies for processing large datasets, commercially known as Big Data. To achieve this goal, software infrastructures (Apache Spark, Apache Mesos, Kubernetes) are employed to optimize the utilization of available computing resources, resulting in a reduction of several orders of magnitude in data processing time. Additionally, the group is involved in developing innovative techniques for real-time (online) acquisition and processing of the enormous amount of signals directly from experiment sensors through the integration of software systems capable of fast data streaming (based on Apache Kafka) towards computing clusters based on Apache Spark.
The resources of the CloudVeneto infrastructure are used in both of these activities through the creation of dedicated Apache Spark/Mesos and Kubernetes clusters.
Dr. Jacopo Pazzini
The project we are carrying out using the CloudVeneto platform involves simulating the recognition process between chemical compounds and target proteins through a molecular docking approach. Currently, in our laboratory (MMS), we have virtually archived a chemical library of approximately 5 million compounds commonly used in screening for identifying new drug candidates.
The archive, called MMsINC, is in the public domain. During the molecular docking simulation, typically 5 plausible ligand-protein complexes are produced for each selected target protein, resulting in a maximum total of approximately 25 million complexes per case study. In our laboratory, this screening campaign is managed through the distribution of various processes.
We are also planning to launch a web service, which we have already named MMSDockCloud, to serve as an access portal to the computation service described above.
Prof. Stefano Moro

CloudVeneto © 2023 – All rights reserved